Tissue images and MS data were provided courtesy of Jessica Moore, Mass Spectrometry Research Center, Department of Chemistry, Vanderbilt University School of Medicine, Nashville, TN, USA.
Application & Background
Biofilms are multicellular microbial communities where cells stick to each other and often adhere to a surface (1). The study of biofilms is important given that most infections in the human body are caused by biofilms originated from one, or a combination of bacterial pathogens (1). Biofilms exhibit tissue-like heterogeneity due to not only the different bacterial subpopulations, but also the spatially-defined differentiation of cells that supports the overall survival of the biofilm (2, 3). During the process of infection, the concentration and distribution of nutrient metals fluctuate due to the efforts by the host to sequester essential minerals to limit pathogenicity in a process known as nutritional immunity (4).
Identification of a particular bacterial subpopulation within a biofilm has been previously achieved by RNA fluorescence in situ hybridization (FISH) and reporter gene fusion techniques (5, 6). The biggest limitation of these methods, however, is that they only support targeted experiments, requiring prior knowledge of the molecule of interest. Furthermore, the number of targets that can be assessed in a single experiment is limited. MALDI mass spectrometry imaging (MALDI MSI) has proven a powerful technology to detect analytes in tissue while preserving their spatial distribution, and thus comprises a unique approach to study bacterial biofilm heterogeneity.
Figure 1. P. aeruginosa biofilms pre- and post-wash.
P. aeruginosa is a biofilm-forming bacterium associated with pulmonary infections, particularly in patients with cystic fibrosis (CF). P. aeruginosa infects the respiratory tracts of 60-70% of CF patients by the age of 20 (7). Here we aimed to study biofilm architecture by visualizing protein distributions within P. aeruginosa biofilms grown both in vitro and developed in CF human lung. In addition, the effect of calprotectin, a key player in nutritional immunity, on biologically-relevant protein profiles was examined.
Figure 2. Summarized experimental work-flow for MSI of bacterial biofilms.
ExperImental
workflow for in vitro grown biofilms
Bacterial biofilms were grown over 5 – 6 days, in a Drip Flow Biofilm Reactor (DFR) (BioSurface Technologies) using glass microscope slides (VWR, USA; #48300-025) as growth surfaces. Figure 2 depicts a diagram of the experimental workflow, from biofilm culture, to MSI data acquisition. Biofilm sections were washed with 70% EtOH for 30 sec, 90% EtOH for 30 sec, 95% EtOH for 30 sec. 3:1 CHCA:DHB matrix was applied in 90% ACN using an HTX TM-Sprayer as described in Table 1. MS imaging of biofilms were performed at 50 µm step size, in linear positive ion mode, with 50 x 50 µm pixels and laser on single beam mode. 50 laser shots were collected in random-walk mode at each pixel.
workflow for biofilms on cystic fibrosis explants
Biopsies from explanted CF lungs were mounted onto chilled ITO coated glass. Tissue was sequentially washed with 70% EtOH (30s), 100% EtOH (30s), 6:2:1 EtOH / chloroform / acetic acid (2min), 100% EtOH (30s), H2O (30s), and EtOH 100% (30s). 3:1 CHCA:DHB matrix was applied in 90% ACN using an HTX TM-Sprayer as described in Table 2. The MALDI MS was operated at 50 by 50mm pixels with the laser in single beam mode. 500 laser shots were collected per pixel, in 50 shot increments.
Table 1. Spray parameters for the in vitro grown biofilms.
Table 2. Spray parameters for the biofilms on the cystic fibrosis explants.
Figure 3. (A) H&E histology of CF human lung (right lower lobe). The inflamed airspace is shown enlarged (inset, gram stained). Optical images obtained at x 20 magnification using a Leica SCN400 Brightfield Slide Scanner. (B) CF human lung at 50 µM using the RapifleX MALDI Tissuetyper, on linear positive ion mode. Overlay: m/z 4,132.222 (green), m/z 4,048.831 (pink), m/z 2,534 (orange), and m/z 2,451 (blue). 100 shots/pixel in 20 µm steps, 4537 positions.
RESULTS
Figure 3A shows an H&E stain of CF human lung tissue presenting significant inflammation. Gram-staining further revealed that bacteria with features consistent with P. aeruginosa and S. aureus co-colonized the infected airspace in the lung. A series of m/z protein ions were observed to localize in the inflamed airspace (Figure 3B). Calprotectin was also found to co-localize in these areas (8).from a CF patient. Selected protein ion images collected with ultra-high speed MALDI-TOF IMS are shown in Figure 3B.
Figure 4A shows MALDI MSI of four different cross-sections of a control P. aeruginosa biofilm. MALDI images highlight the heterogeneity of m/z spatial distributions along, and within, the biofilm. Analysis of P. aeruginosa biofilms challenged with the host protein calprotectin an antimicrobial protein that chelates essential nutrient metals) revealed metal-sensitive bacterial subpopulations within the biomass (Figure 4B).
Figure 4. Four different cross-sections of Pseudomonas aeruginosa strain PA14 biofilms, untreated (A) or exposed to calprotectin in the media (B). Gram-Safranin stained images (post-analysis) are presented on the left, and MALDI MSI signals with differential biofilm localization are shown on the right. Overlay: m/z 6051 (green), m/z 5618 (blue), m/z 9099 (red). MALDI images collected at 50 µm, 66,571 pixels.
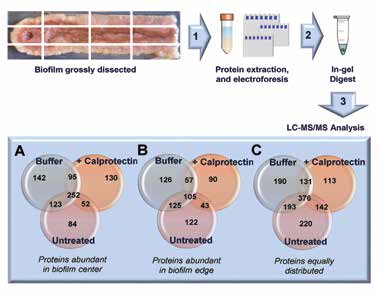
Bottom-up proteomics analysis of grossly-dissected biofilms allowed for the identification of hundreds of proteins related to metabolism, stress, protein synthesis, and other biological processes. MALDI MSI signals identified using bottom-up proteomics (Figure 5) revealed distinct protein profiles in the center compared to the edge of the biofilm, as predicted by the presence of nutrient gradients (i.e. proximal vs. distal to nutrient influx).
These protein profiles were also found responsive to calprotectin exposure (Figure 5A-C). The application of MALDI MSI to the study of structural microbiology significantly furthers our understanding of how microbial communities respond to the host-imposed nutrient limitations during the process of infection.
Figure 5. Summarized work-flow for proteomics analysis of PA14 biofilms. Central channel (A) or nutrient deplete (edge) regions (B) were untreated / treated with buffer / or exposed to calprotectin, dissected and lysed (80% ACN, 5% formic acid, 400 µL bacterial protein extraction reagent) for protein extraction. Protein samples were separated in a 10% Novex Bis-Tris gel at 200V, 5 min. Stained gel bands were excised and subjected to in-gel reduction, alkylation and tryptic digestion. Peptides were sequenced on a Thermo LTQ MS and the resulting spectra were searched against the Uniprot PA14 database using Sequest. Data were compiled using Scaffold version 4.4.3 (8). (A) displays the number of proteins abundant in the biofilm center, (B) in biofilm edges, and (C) proteins that were equally distributed through the biofilm.
CONCLUSIONS
The combination of MALDI MSI, LA-ICP IMS and proteomics provides a powerful tool for the study of protein profiles (i.e. identification and spatial localization) and metal homeostasis within a bacterial biofilm, granting us the opportunity to re-discover biofilm architecture and heterogeneity, which seems tightly correlated with patterns of nutrient(s) availability. The information and data collected by these technologies has high clinical implications given the frequent occurrence of bacterial co-infections in human disease.
References
(1) Peters, B. M., Jabra-Rizk, M. A., O’May, G. A., Costerton, J. W. & Shirtliff, M. E. Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev. 25, 193–213 (2012).
(2) Boles, B. R., Thoendel, M. & Singh, P. K. Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl Acad. Sci. USA 101, 16630–16635 (2004).
(3) Brockhurst, M. A., Hochberg, M. E., Bell, T. & Buckling, A. Character displacement promotes cooperation in bacterial biofilms. Curr. Biol. 16, 2030–2034 (2006).
(4) Hood, M. I. & Skaar, E. P. Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10, 525–537 (2012).
(5) Vlamakis, H., Aguilar, C., Losick, R. & Kolter, R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 22, 945–953 (2008).
(6) Kofoed, M. V., Nielsen, D. A., Revsbech, N. P. & Schramm, A. Fluorescence in situ hybridization (FISH) detection of nitrite reductase transcripts (nirS mRNA) in Pseudomonas stutzeri biofilms relative to a microscale oxygen gradient. Syst. Appl. Microbiol. 35, 513–517 (2012).
(7) Sagel, S. D. et al. Impact of Pseudomonas and Staphylococcus infection on inflammation and clinical status in young children with cystic fibrosis. J. Pediatr. 154, 183–188 (2009).
(8) Wakeman, C. A., Moore, J. L., Noto, M. J., Zhang, Y., Singleton, M. D., Prentice, B. M., Gilston, B. A., Doster, R. S., Gaddy, J. A., Chazin, W. J., Caprioli, R. M.. and Skaar, E. P., The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat. Comm. 7:11951.