The tissue images and MS data presented in this note were provided by Dr. Dusan Velickovic and Dr. Christopher Anderton from Pacific Northwest National Laboratory (PNNL).
Application & Background
Visualizing microbial interactions with matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI) has provided many insights into important biological processes, including metabolic exchange, antibiotic resistance, and microbial competition (1,2). As the application of MALDI-MSI technology to microbial science continues to grow, it is important that validated protocols are shared and established in the literature. Here, we detail a complete experimental workflow for MALDI-MSI of microbial cultures grown on agar, wherein we investigate the different ecological roles of members of a peatland environment.
Peatland ecosystems occupy 3% of the Earth’s land surface yet store 25-33% of terrestrial carbon as recalcitrant organic matter. In the peatland of Marcell experimental forest in northern Minnesota, the ecosystem is dominated by the moss (Sphagnum fallax), which associates with nitrogen-fixing cyanobacteria (Nostoc muscorum) and the fungus (Trizodia spp.), but these interactions are not well characterized. In this study, under the mission of the Spruce-Peatland Response Under Climate and Environmental Change (SPRUCE) Department of Energy (DOE), we aimed to identify the role of various peatland species and to investigate the metabolic communication between them in order to better understand and control biodiversity within this complex ecosystem (3).
Experimental
SAMPLE PREPARATION
For tripartite interactions, each organism was positioned approximately 2 cm from one another and for individual control, a single organism was placed in the center of the plate (2). All samples to be analyzed by MALDI IMS were grown on 1.5 mm thick BG-110 (pH 5.5) 1.5% agar plates at 24°C with a 16 hr/8 hr (day/night) cycle at 150 PAR. The petri dishes were sealed with Parafilm to minimize dehydration of thin agar during incubation and incubated for 12 days. After the incubation period, agar areas of the tripartite interaction and of isolated culture controls were excised from petri dishes and were carefully placed onto an indium tin oxide (ITO)-coated glass slide using a spatula. Since the moss excessively protruded from the surface of the agar, plant tissue was carefully removed prior to dehydration to assist in MSI analyses. Samples were then dried under ambient conditions in a BSL hood overnight (2).
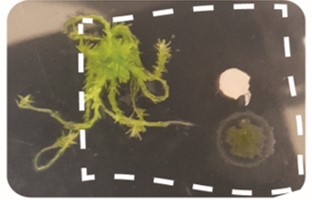
Figure 1A. The tripartitre interation of the moss (Sphagnum fallax), nitrogen-fixing bacteria (Nostoc muscorum), and the fungus (Trizodia spp.) grown in a petri dish on 1.5% BG110 agar.
Figure 1B. The agar area of the tripartite interaction was excised from the petri dish and transferred onto an ITO-coated glass slide. Samples were dehydrated overnight at ambient temperature and ambient pressure before DHB MALDI matrix was applied using the HTX TM-Sprayer.
Matrix Deposition
CHCA was applied to the slides at a concentration of 10 mg/mL (in 50:50 acetonitrile:water) using the HTX TM-Sprayer Sprayer (3). The spraying took 12 minutes per slide (25x75mm). The slides were coated using the following parameters:
Table 1. Spray parameters for MALDI matrix deposition.
MALDI Mass Spectrometry Imaging
Imaging was performed on a 15 Tesla MALDI FTICR-MS (SolariX, Bruker Daltonics) equipped with a SmartBeam II laser source (355 nm, 2 kHz) in positive mode, using 200 shots at 2kHz and a 200 μm step size. The SolariX FTICR-MS was operated to collect m/z 92-1000, using a 209 ms transient, which translated to a mass resolution of R ~ 130,000 at m/z 400. Data was acquired using FlexImaging (v4.1, Bruker Daltonics), and imaging processing and visualization were performed using SCiLS Lab.
Results
Using spatial segmentation image analysis in SCiLS, 288 ion images associate with members of tripartite community (Figure 2). For 213 ion images, molecular formulas are assigned based on high accurate m/z. These data are also uploaded in METASPACE, and are publicly available for browsing here.*
As seen in Figure 3, adenine was found to co-localize with the cyanobacteria. This was an expected finding as adenine is known to play an important role in maintaining energy balance during the nitrogen fixation process (4). Butyrylcarnitine, described as a signaling molecule in plant-fungus symbiosis, was almost exclusively mapped around the fungi (5). Finally, choline-sulfate, a plant osmoprotectant, showed co-localization with the plant part of the tripartite community (6).
Figure 2. SCiLS spectral segmentation of tripartite interaction
Figure 3. Distribution of some metabolites involved in tripartite communication. Lateral resolution: 200 μm
Conclusions
In MALDI IMS experiments involving complex systems composed from several species, tracking spatial metabolism of specific compounds can be an efficient solution for the structural identification of these compounds. In this study, the ion m/z 189.1598 was identified as [C9H20N2O2 + H]+ and was distributed around the fungus in the tripartite interaction. This ion can be ascribed to two previously identified natural compounds: 7,8-diaminononanoate and trimethyllysine, metabolites with related, but distinct biological roles in fatty acid metabolism (Figure 4). Specifically, trimethyllysine is precursor of carnitine biosynthesis, a molecule that serves as a fatty acid shuttle during lipid catabolism, while 7,8-diaminononanoate plays a role in the biosynthesis of biotin, an essential carbon dioxide carrier co-factor for fatty acid synthesis (7,8). When we imaged the end-products of these two biosynthetic routes we can see much higher co-localization (Pearson correlation coefficient 0.7) of biotin amide than carnitine (Pearson correlation coefficient 0.0) with [C9H20N2O2 + H]+ ion. This indicate that observed ion is rather 7,8-diaminonanoate than trimethyllisine. Note that MS/MS data are not able to unambiguously resolve these two isomers, so spatial metabolomics could be advanced approach in such cases (2).
Figure 4. Tracking the metabolic routes of possible candidates of ion m/z 189.1598 in a spatial manner in order to identify which of two natural compounds it represents. Co-localization of ion m/z 266.0934 and m/z 189.1598 and absence of any co-localization between m/z 162.1125 and m/z 189.1598 indicate that the ion m/z 189.1598 with the molecular formula C9H20N2O2 is 7,8 diaminononaoate. Lateral resolution: 200 μm.
REFERENCES
(1) Anderton, C. R., Chu, R. K.,Tolic, N., Creissen, A. and Pasa-Tolic, L. (2016). "Utilizing a Robotic Sprayer for High Lateral and Mass Resolution MALDI FT-ICR MSI of Microbial Cultures." Journal of the American Society for Mass Spectrometry 27(3): 556-559.
(2) Velickovic, D., Chu, R.K., Carrell, A.A., Thomas, M., Pasa-Tolic, L., Weston, D. J. and Anderton, C. R. (2018). "Multimodal MSI in Conjunction with Broad Coverage Spatially Resolved MS2 Increases Confidence in Both Molecular Identification and Localization." Analytical Chemistry 90(1): 702-707.
(3) Hanson, P. J., Childs, K. W., Wullschleger, S. D., Riggs, J. S., Thomas, W.K., Todd , D. E. and Warren, J. M. (2011). "A method for experimental heating of intact soil profiles for application to climate change experiments." Global Change Biology 17(2): 1083-1096.).
(4) Privalle, L. S. and Burris, R. H. (1983). "Adenine-Nucleotide Levels in and Nitrogen-Fixation by the Cyanobacterium Anabaena Sp Strain-7120." Journal of Bacteriology 154(1): 351-355.
(5) Laparre, J., Malbreil, M., Letisse, F., Portais, J. C., Roux, C., Becard, G., and Puech-Pages, V. (2014). "Combining Metabolomics and Gene Expression Analysis Reveals that Propionyl- and Butyryl-Carnitines Are Involved in Late Stages of Arbuscular Mycorrhizal Symbiosis." Molecular Plant 7(3): 554-566.
(6) Rivoal, J. and A. D. Hanson (1994). "Choline-O-Sulfate Biosynthesis in Plants - Identification and Partial Characterization of a Salinity-Inducible Choline Sulfotransferase from Species of Limonium (Plumbaginaceae)." Plant Physiology 106(3): 1187-1193.
(7) Franken, J., Burger, A., Swiegers, J. H., and Bauer, F. F. (2015). "Reconstruction of the carnitine biosynthesis pathway from Neurospora crassa in the yeast Saccharomyces cerevisiae." Applied Microbiology and Biotechnology 99(15): 6377-6389.
(8) Wang, M. L., Moynie, L.,Harrison, P. J., Kelly, V., Piper, A., Naismith, J. H. and Campopiano, D. J. (2017). "Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters." Nature Chemical Biology 13(6): 660-+.